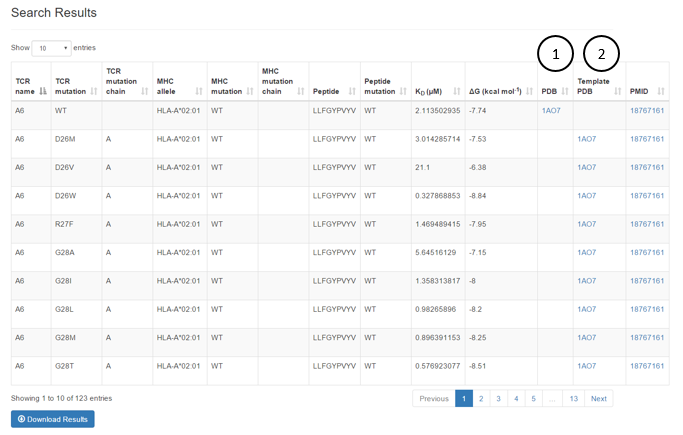
Search Example
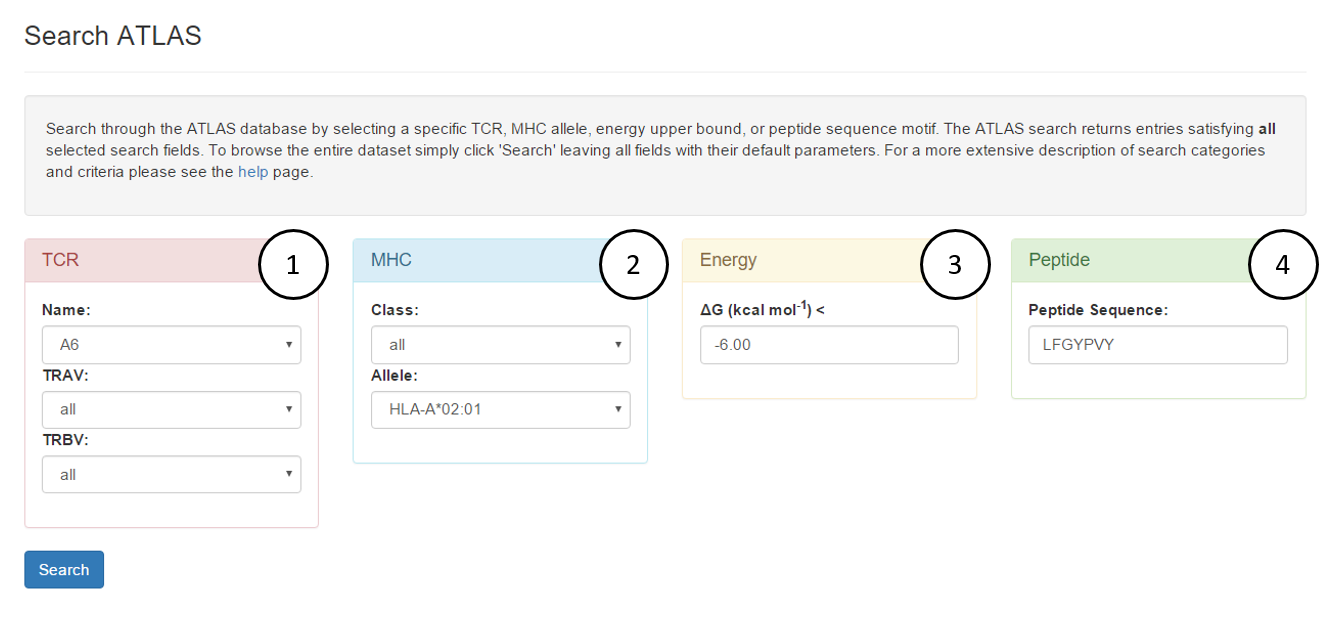
-

Search by TCR name or by T Cell receptor alpha/beta variable genes (TRAV/TRBV)
Name: A6
-

Search by MHC allele or view all entries for an MHC class
Allele: HLA-A*02:01
-

Search for entries with experimental binding energies below input ΔG in kcal mol-1
ΔG (kcal mol-1) < -6.00
-

Search for entries containing a peptide with input amino acid sequence. Sub-sequences are allowed. Case-insensitive.
Peptide sequence: LFGYPVY